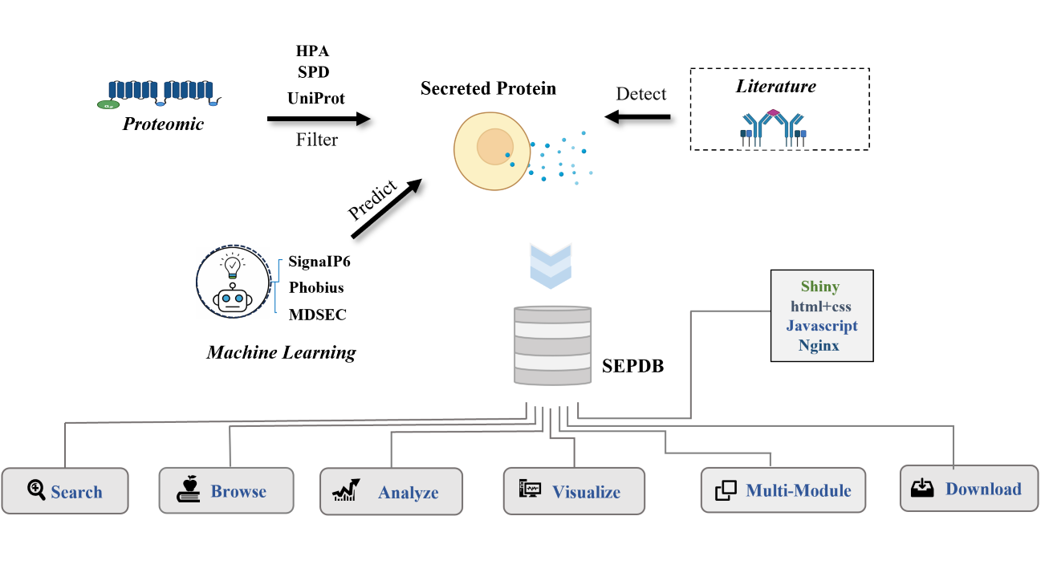
SEPDB collected verified data from the existing protein database, collected human secreted proteomic HSPA through
N-terminal signal peptide prediction and transmembrane domain signal peptide verification, and collected other
Rat/Mouse secretory protein comparison, through the artificially collected secretory data was labeled to prove the
specific information of secreted proteins, users can not only check the secretion in time The evidence for the protein,
and the prediction of the unknown protein, when compared with the basic experiment, the publicly secreted protein
will be more convincing and more extensive.
Overview of secreted protein datasets
SEPDB integrates datasets from the literature (e.g., serum, exosomes, and tissue cultures), subcellular localization datasets from a priori databases, and datasets calculated by models. Organize modules for aging-related secreted protein datasets and exercise-related secreted protein datasets.
SEPDB integrates existing human secretory proteomics with N-terminal signal peptide prediction and transmembrane structural domain signal peptide validation to create the human secretory proteomics. We have supplemented the dataset with rat and mouse biological validation data from different models.
SEPDB is an integrated database with secretory proteomics
that provides human, mouse,and rat secretory proteomics
datasets by collecting data in serum, exosomes, and cell
culture media.
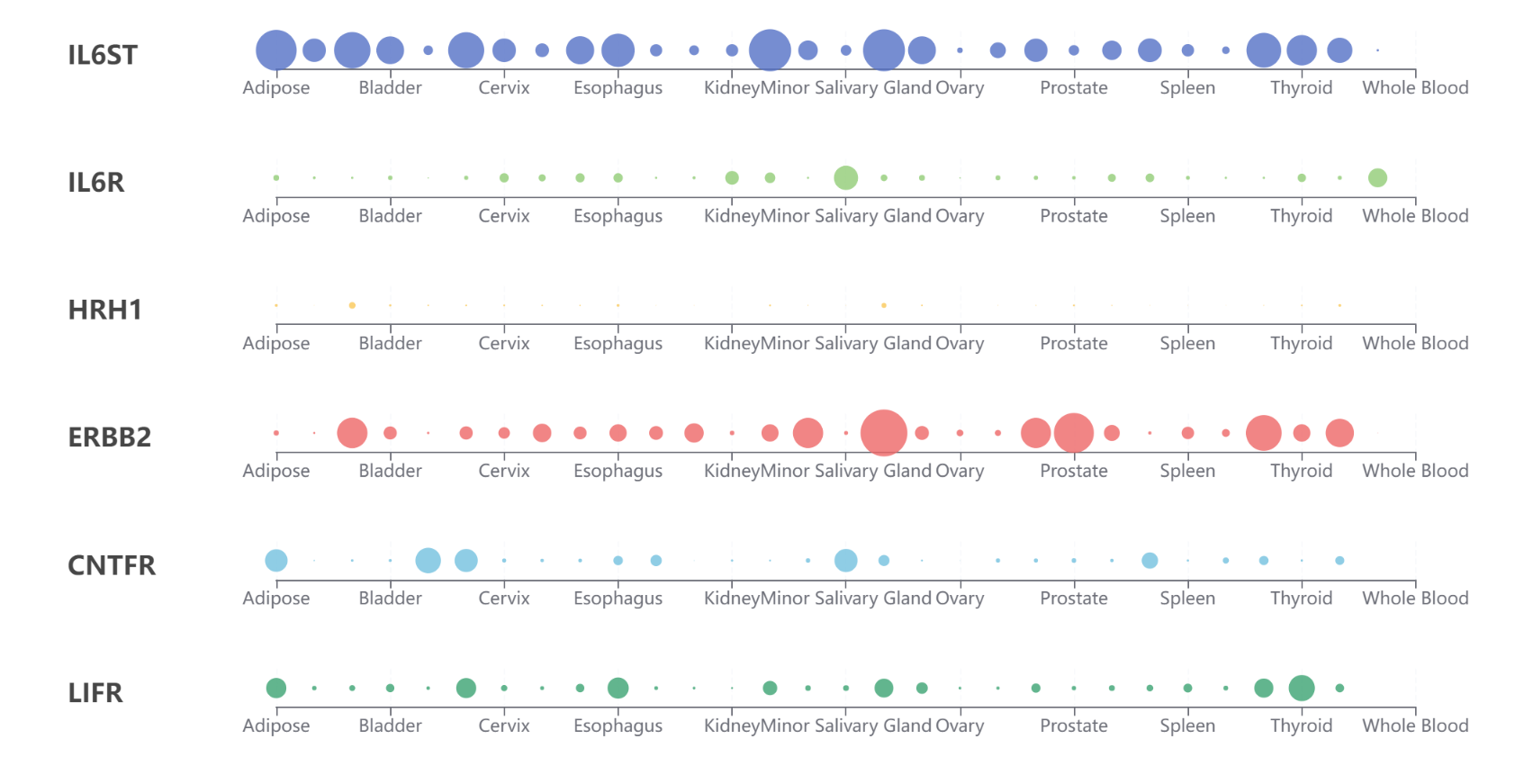
Tissue specific expression network of IL6 receptor
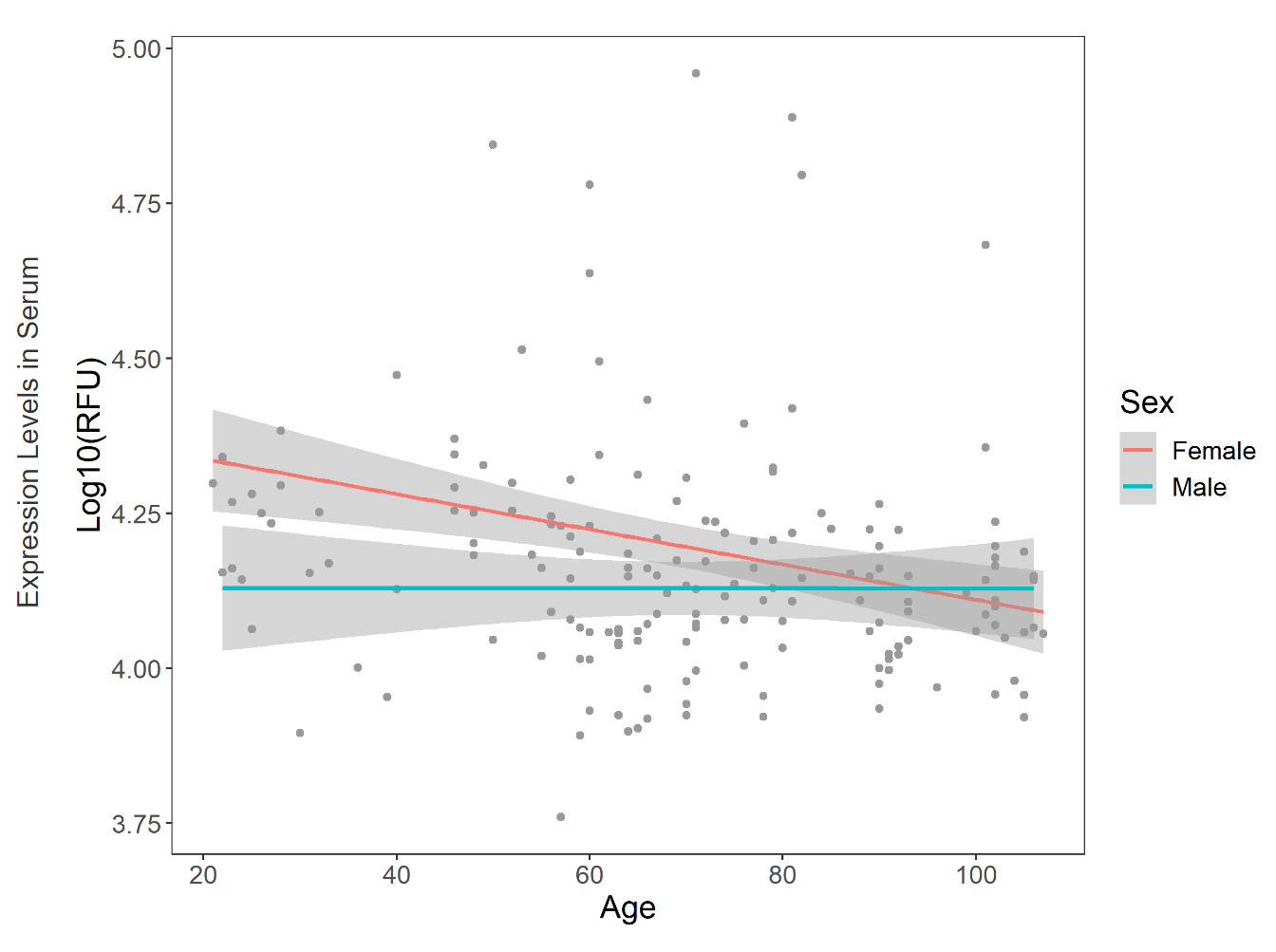
Expression changes of A2M search gene senescence related secreted protein in 170 samples

SEPDB integrates a pair of secretory protein cell types, some of which are related to movement.